

Contains the writeOutput() method for saving output files to disk and the writeInput() method for saving input files to disk. A number of helper functions are used by both functions due to the high degree of similarity between the input and output file syntax.
Write a MEASURE input file to path on disk. The parameters needed mirror those returned by readInput():
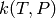
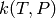 values to
values toIf successful, the file created on disk should be able to be read in by readInput() with (hopefully) no loss of fidelity.
Write a MEASURE output file to path on disk. The parameters needed mirror those returned by readInput():
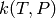
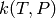 values to
values toIf successful, the file created on disk will contain all of the species
and net reaction data, including all phenomenological rate coefficients
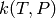 .
.
Write all species in the given unimolecular reaction network to a file object`f`. All isomer, reactant, product, and bath gas species are automatically written one time each.
Write all path reactions in the given unimolecular reaction network to a file object`f`. The path reactions are those reactions that directly connect adjacent molecular configurations; these are the reactions that remain in the high-pressure limit.
Write all net reactions in the given unimolecular reaction network to a file object`f`. The net reactions are those reactions that can connect any pair of molecular configurations, not just those directly adjacent. These are the reactions that have pressure-dependent rate coefficients.
Write a Species object spec to a file object f.
Write a Reaction object rxn to a file object f.
Write a Reaction object rxn that has pressure-dependent kinetics to a file object f.
Write the StatesModel data states containing molecular degree of freedom data to the file object f. The optional parameter prefix is prepended to each line of the output file, which provides an easy way to adjust the indentation.